OPTIMIZATION OF ANNEALING TEMPERATURE AND PRIMER CONCENTRATION OF CYTOCHROME B (CYT B) GENE FOR PIG DNA DETECTION WITH REAL-TIME PCR METHOD
Downloads
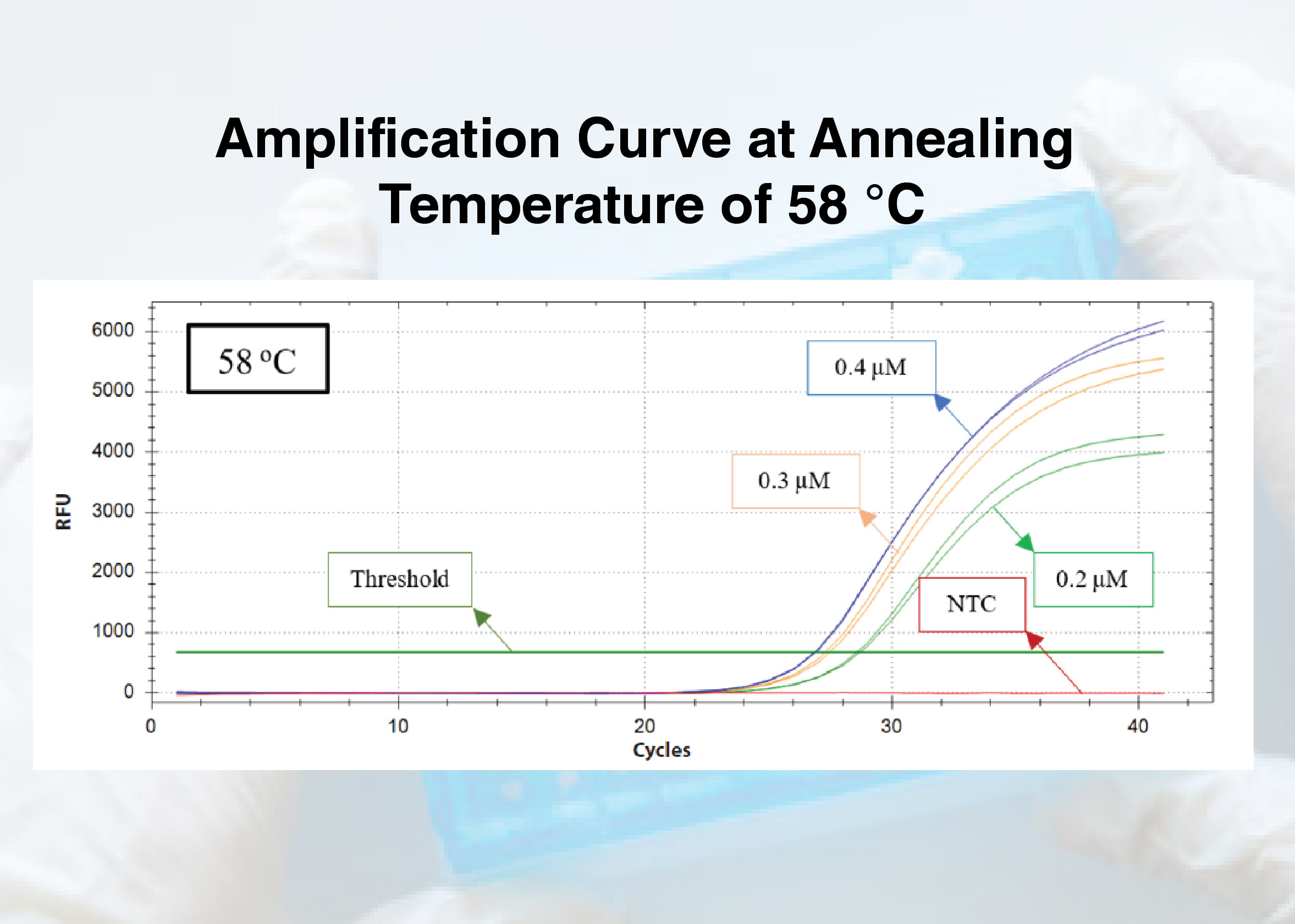
Background: One company in Indonesia has developed a pig DNA detection kit by designing primers with the real-time Polymerase Chain Reaction (PCR) method using the cytochrome b (cyt b) gene. It is necessary to optimize the PCR process to optimize pig DNA detection, including annealing temperature and primer concentration, which can increase sensitivity, specificity, and precision. Purpose: This research aims to determine the optimum annealing temperature and primer concentration for the detection of pig DNA using cyt b gene. Method: In this research, the extracted sample isolates were subjected to 12 treatments with 2 repetitions. Optimal data analysis was based on the lowest Cycle Threshold (CT) value in the amplification curve. Result: Out of a total of 24 samples, an increase in the CT value was observed at annealing temperatures of 57 °C, 59 °C, and 60 °C compared to 58 °C, across various primer concentrations. The primer concentrations with the lowest CT values were successively found to be 0.4 μM, 0.3 μM, and 0.2 μM. Conclusion: The results of the research that has been conducted indicate that the optimal annealing temperature for detecting pig DNA using the cyt b gene in this research is 58 °C, and the optimal concentrations of forward and reverse primers are 0.4 μM.
Amaral, J.S., Santos, G., Oliveira, M.B.P.P., Mafra, I., 2017. Quantitative Detection of Pork Meat by Evagreen Real-Time PCR to Assess The Authenticity of Processed Meat Products. Food Control Vol. 72, Pp. 53–61.
Arya, M., Shergill, I.S., Williamson, M., Gommersall, L., Arya, N., Patel, H.R.H., 2005. Basic Principles of Real-Time Quantitative PCR. Expert Review of Molecular Diagnostics Vol. 5(2), Pp. 209-219.
BIO-RAD Laboratories, 2006. Real-Time PCR Applications Guide.
BIO-RAD Laboratories, 2022. CFX96 Touch Real-Time PCR Detection System | Bio-Rad. BIO-RAD. URL https://www.bio-rad.com/en-id/product/cfx96-touch-real-time-pcr-detection-system?ID=LJB1YU15 (accessed 2.1.24).
Borah, P., 2011. Primer Designing for PCR. Science Vision Vol. 11(3), Pp. 134-136.
Bustin, S.A., Benes, V., Garson, J.A., Hellemans, J., Huggett, J., Kubista, M., Mueller, R., Nolan, T., Pfaffl, M.W., Shipley, G.L., Vandesompele, J., Wittwer, C.T., 2009. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clinical Chemistry Vol. 55(4), Pp. 611-622.
Caraguel, C.G.B., Stryhn, H., Gagné, N., Dohoo, I.R., Hammell, K.L., 2011. Selection of A Cut Off Value for Real-Time Polymerase Chain Reaction Results to Fit A Diagnostic Purpose: Analytical and Epidemiologic Approaches. Journal of Veterinary Diagnostic Investigation: Official Publication of The American Association of Veterinary Laboratory Diagnosticians, Inc Vol. 23(1), Pp. 2-15.
Dooley, J.J., Paine, K.E., Garrett, S.D., Brown, H.M., 2004. Detection of Meat Species using Taqman Real-Time PCR Assays. Meat Science Vol. 68(3), Pp. 431-438.
Farias, I.P., Ortí, G., Sampaio, I., Schneider, H., Meyer, A., 2001. The Cytochrome b Gene as A Phylogenetic Marker: The Limits of Resolution for Analyzing Relationships among Cichlid Fishes. Journal of Molecular Evolution. Journal of Molecular Evolution Vol. 53(2), Pp. 89-103.
Gunson, R., Gillespie, G., F Carman, W., 2003. Optimisation of PCR Reactions using Primer Chessboarding. Journal of Clinical Virology Vol. 26(3), Pp. 369-373.
Handoyo, D., Rudiretna, A., 2000. Prinsip Umum dan Pelaksanaan Polymerase Chain Reaction (PCR). UNITAS Vol. 9(1), Pp. 17-29.
Harshitha, R., Arunraj, D.R., 2021. Real‐Time Quantitative PCR : A Tool for Absolute and Relative Quantification. Biochemistry and Molecular Biology Education. Biochemistry and Molecular Biology Education: A Bimonthly Publication of the International Union of Biochemistry and Molecular Biology Vol. 49(5), Pp. 800-812.
Hilscher, C., Vahrson, W., Dittmer, D.P., 2005. Faster Quantitative Real-Time PCR Protocols May Lose Sensitivity and Ahow Increased Variability. Nucleic Acids Research Vol. 33(21), Pp. e182.
Hsu, J.T., Das, S., Mohapatra, S., 1997. Polymerase Chain Reaction Engineering. Biotechnology and Bioengineering Vol. 55(2), Pp. 359-366.
Hudson, 2008. Hudson Genomics. Hudson Institute of Medical Research. URL https://www.hudson.org.au/facilities/hudson-genomics/ (accessed 2.1.24).
IDT, 2022. Primer Design Tools for PCR & Amp; qPCR. Integrated DNA Technologies. URL https://sg.idtdna.com/pages/education/decoded/article/design-efficient-pcr-and-qpcr-primers-and-probes-using-online-tools (accessed 2.1.24).
Janudin, A., Chin, N., Ahmed, M., 2022. An Eva Green Real-Time PCR Assay for Porcine DNA Analysis in Raw and Processed Foods. Malaysian Journal of Halal Research Vol. 5(1), Pp. 33039.
Joko, T., Kusumandari, N., Hartono, S., 2011. Optimasi Metode PCR untuk Deteksi Pectobacterium carotovorum, Penyebab Penyakit Busuk Lunak Anggrek. Jurnal Perlindungan Tanaman Indonesia Vol. 17(2), Pp. 54-59.
Kim, Y.S., Yu, H.K., Lee, B.Z., Hong, K.W., 2018. Effect of DNA Extraction Methods on The Detection of Porcine Ingredients in Halal Cosmetics using Real-Time PCR. Applied Biological Chemistry Vol. 61(5), Pp. 549-555.
Life Technologies, 2014. Real-Time PCR Handbook.
Markoulatos, P., Siafakas, N., Moncany, M., 2002. Multiplex Polymerase Chain Reaction: A Practical Approach. Journal of Clinical Laboratory Analysis Vol. 16(1), Pp. 47-51.
Ministry of Religion, 2014. Jaminan Produk Halal.
Pestana, E.A., Belák, S., Diallo, A., Crowther, J., Viljoen, G., 2010. Early, Rapid and Sensitive Veterinary Molecular Diagnostics-Real Time PCR Applications. Springer, Dordrecht Heidelberg, London.
Republika, 2021. Pengajuan Sertifikasi Halal Meningkat, Ini Alasannya.
Roux, K.H., 2009. Optimizing and Troubleshooting in PCR. Cold Spring Harbor Protocols Vol. 2009(4), Pp. pdbip66.
Tania, Nolan, Bergkvist, A., Carvallo, C., Chereson, P.H., Daley, L., Heath, A., Hibb, S., Hoge, S., Jouravlena, E., Kreader, C., Mohammed, M., Mueller, E., Richardson, G., Russell, T., Ward, B., Weber, S.A., Wiklander, M., 2014. A Technical Guide PCR Technologies. SIGMA-AL
Copyright (c) 2025 Journal of Vocational Health Studies

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
- The authors agree to transfer the transfer copyright of the article to the Journal of Vocational Health Studies (JVHS) effective if and when the paper is accepted for publication.
- Legal formal aspect of journal publication accessibility refers to Creative Commons Attribution-NonCommercial-ShareAlike (CC BY-NC-SA), implies that publication can be used for non-commercial purposes in its original form.
- Every publications (printed/electronic) are open access for educational purposes, research, and library. Other that the aims mentioned above, editorial board is not responsible for copyright violation.
Journal of Vocational Health Studies is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License