Date Log
Copyright (c) 2023 Jurnal Ilmiah Perikanan dan Kelautan

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
1. Copyright of the article is transferred to the journal, by the knowledge of the author, whilst the moral right of the publication belongs to the author.
2. The legal formal aspect of journal publication accessibility refers to Creative Commons Atribusi-Non Commercial-Share alike (CC BY-NC-SA), (https://creativecommons.org/licenses/by-nc-sa/4.0/)
3. The articles published in the journal are open access and can be used for non-commercial purposes. Other than the aims mentioned above, the editorial board is not responsible for copyright violation
The manuscript authentic and copyright statement submission can be downloaded ON THIS FORM.
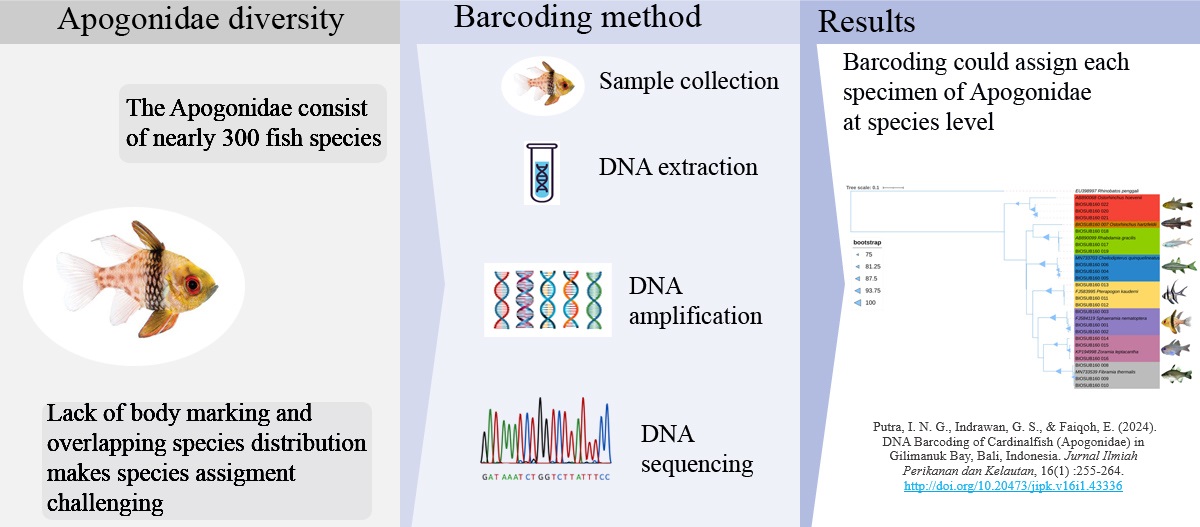
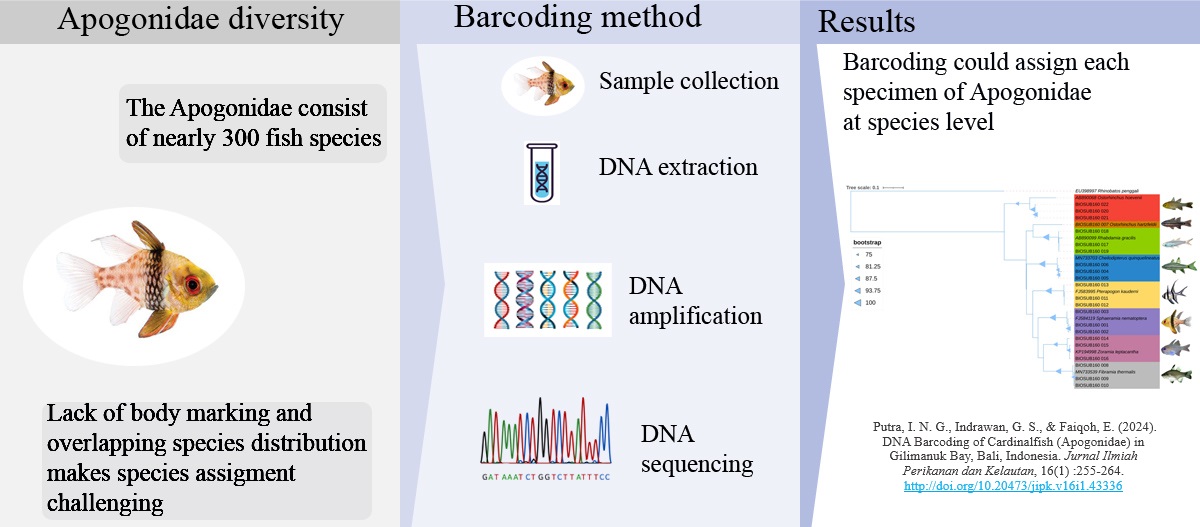
DNA Barcoding of Cardinalfish (Apogonidae) in Gilimanuk Bay, Bali, Indonesia
Corresponding Author(s) : I Nyoman Giri Putra Putra
Jurnal Ilmiah Perikanan dan Kelautan, Vol. 16 No. 1 (2024): JURNAL ILMIAH PERIKANAN DAN KELAUTAN
Abstract
Abstract
The Apogonidae is estimated to consist of nearly 300 fish species, most of which inhabit coral reef areas. The lack of distinctive body markings and overlapping species distribution makes species assignment challenging. Therefore, this study aimed to delineate species and establish barcoding reference databases of Apogonidae in Gilimanuk Bay (Bali, Indonesia) using the Cytochrome Oxidase I (COI) gene of the mitochondrial DNA. A total of 22 fish tissue samples were extracted with 10% Chelex solution. BLAST analysis was performed and genetic differentiation between species was calculated. The phylogenetic tree was constructed using the Maximum Likelihood method and tree visualization was generated using iTOL V5. The morphology and genetic identification results based on the mitochondrial COI gene revealed eight species of seven genera, and one species was new to GenBank online database. This study was the first-ever addition of COI sequence for Ostorhinchus hartzfeldii into the GenBank database. The average K2P genetic distance within species and K2P distance between genera within the family were 0.60% and 19.10%, respectively. The mean genetic distance between genera within the family was 31.8-fold higher than the mean genetic distance within species. The phylogenetic tree showed that each sample resided in a distinct cluster, which indicates that DNA barcoding is a reliable and effective approach for species delimitation in Apogonidae fishes.
Highlight Research
- Eight species of cardinalfish in Gilimanuk Bay were delineated using the barcoding method.
- This study provides the first nucleotide sequence for Ostorhinchus hartzfeldii in GenBank online database.
- The phylogenetic tree showed that each sample resided in a distinct cluster, indicating that the barcoding method efficiently differentiated at the species level.
- The mean genetic distance between genera within the family was 31.8-fold higher than the mean genetic distance within species.
Keywords
Download Citation
Endnote/Zotero/Mendeley (RIS)BibTeX
- Abecia, J. E. D., Luiz, O. J., & King, A. J. (2018). Intraspecific morphological and reproductive trait variation in mouth almighty Glossamia aprion (Apogonidae) across different flow environments. Journal of Fish Biology, 93(5):961-971.
- Afiati, N., Subagiyo, Handayani, C. R., Hartati, R., & Kholilah, N. (2022). DNA barcoding on Indian Ocean squid, Uroteuthis duvaucelii (D'Orbigny, 1835) (Cephalopoda: Loliginidae) from the Java Sea, Indonesia. Jurnal Ilmiah Perikanan dan Kelautan, 14(2):231-245.
- Alfonsi, E., Méheust, E., Fuchs, S., Carpentier, F. G., Quillivic, Y., Viricel, A., Hassani, S., & Jung, J. L. (2013). The use of DNA barcoding to monitor the marine mammal biodiversity along the French Atlantic coast. Zookeys, 365:5-24.
- Allen, G. R., & Erdmann, M. V. (2012a). Reef fishes of Bali, Indonesia. In P. L. K. Mustika, I. M. J. Ratha, and S. Purwanto (Ed.), The 2011 Bali marine rapid assessment (Second English edition). (pp. 15-68). Bali: Conservation International.
- Allen, G. R., & Erdmann, M. V. (2012b). Reef Fishes of the East Indies. Volume I-III. Virginia: Conservation International Foundation.
- Alvarenga, M., Solé-Cava, A. M., & Henning, F. (2021). What's in a name? Phylogenetic species identification reveals extensive trade of endangered guitarfishes and sharks. Biological Conservation, 257:109-119.
- Arbi, U. Y., Suharti, S. R., Huwae, R., Rizqi, M. P., & Suratno. (2019). Populasi ikan endemik Capungan Banggai (Pterapogon kauderni) di habitat introduksi di teluk Gilimanuk, Bali. Paper presented at the Seminar Nasional Tahunan XVI Hasil Penelitian Perikanan dan Kelautan, Universitas Gadjah Mada, Indonesia.
- Arroyave, J., & Stiassny, M. L. J. (2014). DNA barcoding reveals novel insights into pterygophagy and prey selection in distichodontid fishes (Characiformes: Distichodontidae). Ecology and Evolution, 4(23):4534-4542.
- Barber, P. H., Erdmann, M. V, & Palumbi, S. R. (2006). Comparative phylogeography of three codistributed stomatopods: origins and timing of regional lineage diversification in the Coral Triangle. Evolution, 60(9):1825-1839.
- Bingpeng, X., Heshan, L., Zhilan, Z., Chunguang, W., Yanguo, W., & Jianjun, W. (2018). DNA barcoding for identification of fish species in the Taiwan Strait. PLoS ONE, 13(6):e0198109.
- Cappenberg, H. A. W., Aznam, A., & Aswandy, I. (2006). Komunitas moluska di perairan Teluk Gilimanuk, Bali Barat. Oseanologi dan Liminologi di Indonesia, 40:53-64.
- Cinner, J. E., & Mcclanahan, T. R. (2006). Socioeconomic factors that lead to overfishing in small-scale coral reef fisheries of Papua New Guinea. Environmental Conservation, 33(1):73-80.
- Delrieu-Trottin, E., Durand, J. D., Limmon, G., Sukmono, T., Kadarusman, Sugeha, H. Y., Chen, W. J., Busson, F., Borsa, P., Dahruddin, H., Sauri, S., Fitriana, Y., Zein, M. S. A., Hocdé, R., Pouyaud, L., Keith, P., Wowor, D., Steinke, D., Hanner, R., & Hubert, N. (2020). Biodiversity inventory of the grey mullets (Actinopterygii: Mugilidae) of the Indo-Australian Archipelago through the iterative use of DNA-based species delimitation and specimen assignment methods. Evolutionary Applications, 13(6):1451-1467.
- Fadli, N., Nor, S. A. M., Othman, A. S., Sofyan, H., & Muchlisin, Z. A. (2020). DNA barcoding of commercially important reef fishes in Weh Island, Aceh, Indonesia. PeerJ, 8(2012):1-25.
- Falah, I. N., Adharini, R. I., & Ratnawati, S. E. (2023). Molecular identification of elvers (Anguilla spp.) from river estuaries in Central Java, Indonesia using DNA barcoding based on mtDNA CO1 sequences. Jurnal Ilmiah Perikanan dan Kelautan, 15(1):121–130.
- Fraser, T. H. (2006). A new species of cardinalfish (Perciformes: Apogonidae: Apogon) from New Caledonia, with comments and a key to related species. Proceedings of the Biological Society of Washington, 119(1):137-142.
- Fraser, T. H., Bogorodsky, S. V., Mal, A. O., & Alpermann, T. J. (2021). Review of the cardinalfishes of the genus Cercamia (Percomorpha: Apogonidae) of the Red Sea and Indian Ocean with descriptions of three new species. Zootaxa, 5039(3):363-394.
- Fraser, T. H., Randall, J. E., & Allen, G. R. (2002). Clarification of the cardinalfishes (Apogonidae) previously confused with Apogon moluccensis valenciennes, with a description of a related new species. The Raffles Bulletin of Zoology, 50(1):175-184.
- Gardiner, N. M., & Jones, G. P. (2005). Habitat specialisation and overlap in a guild of coral reef cardinalfishes (Apogonidae). Marine Ecology Progress Series, 305:163-175.
- Gon, O., & Allen, G. R. (2012). Revision of the Indo-Pacific cardinalfish genus Siphamia (Perciformes: Apogonidae). Zootaxa, 3294(1):1-84.
- Keyse, J., Treml, E. A., Huelsken, T., Barber, P. H., Deboer, T., Kochzius, M., Nuryanto, A., Gardner, J. P. A., Liu, L. L., Penny, S., & Riginos, C. (2018). Historical divergences associated with intermittent land bridges overshadow isolation by larval dispersal in co-distributed species of Tridacna giant clams. Journal of Biogeography, 45(4):848-858.
- Kimura, M. (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 16:111-120.
- Kumar, S., Stecher, G., Li, M., Knyaz, C., & Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Molecular Biology and Evolution, 35(6):1547-1549.
- Landi, M., Dimech, M., Arculeo, M., Biondo, G., Martins, R., Carneiro, M., Carvalho, G. R., Lo Brutto, S., & Costa, F. O. (2014). DNA barcoding for species assignment: The case of Mediterranean marine fishes. PLoS ONE, 9(9):e106135.
- Lazuardi, M. E., Sudiarta, I. K., Ratha, I. M. J., Ampou, E. E., Nugroho, S. C., & Mustika, P. L. (2012). The status of coral reefs in Bali. In P. L. K. Mustika, I. M. J. Ratha, and S. Purwanto (Ed.), The 2011 Bali marine rapid assessment (Second English edition). (pp. 69-77). Bali: Conservation International.
- Lea, R. N., Fraser, T. H., Baldwin, C. C., & Craig, M. T. (2022). Five valid species of Cardinalfishes of the Genus Apogon (Apogonidae) in the Eastern Pacific Ocean, with a redescription of A. atricaudus and notes on the distribution of A. atricaudus and A. atradorsatus. Ichthyology & Herpetology, 110(1):106-114.
- Lessios, H. A., Kane, J., & Robertson, D. R. (2003). Phylogeography of the pantropical sea urchin Tripneustes: contrasting patterns of population structure between oceans. Evolution, 57(9):2026.
- Letunic, I., & Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Research, 49(W1):W293-W296.
- Limmon, G., Delrieu-Trottin, E., Patikawa, J., Rijoly, F., Dahruddin, H., Busson, F., Steinke, D., & Hubert, N. (2020). Assessing species diversity of coral triangle artisanal fisheries: A DNA barcode reference library for the shore fishes retailed at Ambon harbor (Indonesia). Ecology and Evolution, 10(7):3356-3366.
- Luehrmann, M., Cortesi, F., Cheney, K. L., de Busserolles, F., & Marshall, N. J. (2020). Microhabitat partitioning correlates with opsin gene expression in coral reef cardinalfishes (Apogonidae). Functional Ecology, 34(5):1041-1052.
- Ma'ruf, M. S., Arthana, I. W., & Ernawati, N. M. (2022). Composition of types and conditions of mangrove in Gilimanuk Bay, West Bali National Park. Ecotrophic, 16(2):153-173.
- Maas, D. L., Capriati, A., Ahmad, A., Erdmann, M. V., Lamers, M., de Leeuw, C. A., Prins, L., Purwanto, Putri, A. P., Tapilatu, R. F., & Becking, L. E. (2020). Recognizing peripheral ecosystems in marine protected areas: A case study of golden jellyfish lakes in Raja Ampat, Indonesia. Marine Pollution Bulletin, 151:110700.
- Mabuchi, K., Fraser, T. H., Song, H., Azuma, Y., & Nishida, M. (2014). Revision of the systematics of the cardinalfishes (Percomorpha: Apogonidae) based on molecular analyses and comparative reevaluation of morphological characters. Zootaxa, 3846(2):151-203.
- Mbaba, K. Y., Suryatika, I. B. M., & Adnyana, I. G. A. P. (2019). Analysis of the concentration of Cd metal in Rhizophora.Sp using the AAS method in the Gilimanuk Harbor area, Jembrana, Bali. Kappa Journal, 3:83-88.
- Meyers-Muñoz, M. A., van der Velde, G., van der Meij, S. E. T., Stoffels, B. E. M. W., van Alen, T., Tuti, Y., & Hoeksema, B. W. (2016). The phylogenetic position of a new species of Plakobranchus from West Papua, Indonesia (Mollusca, Opisthobranchia, Sacoglossa). Zookeys, 594:73-98.
- Nei, M., & Kumar, S. (2000). Molecular evolution and phylogenetics. New York: Oxford University Press.
- Pamungkas, P. B. P., Hendrawan, I. G., & Putra, I. N. G. (2021). Characteristics and distribution of stranded waste in the coastal area of West Bali National Park. Journal of Marine Research and Technology, 4(1):9-15.
- Permana, G. N., Slamet, B., Permana, B. A., Dewi, A. K., & Mahardika, G. N. (2019). Population genetic structure of spiny lobsters, Panulirus homarus and Panulirus ornatus, in the Indian Ocean , Coral Triangle, and South China Sea. Indonesian Aquaculture Journal, 14(1):7-14.
- Pettaloloa, D., Watiniasih, N., & Sari, A. (2022). The diversity of water bird species in the Karang Sewu mangrove forest, Gilimanuk, Bali. Current Trends in Aquatic Science, 3:45-51.
- Pöppe, J., Sutcliffe, P., Hooper, J. N. A., Wörheide, G., & Erpenbeck, D. (2010). COI Barcoding reveals new clades and radiation patterns of Indo-Pacific sponges of the family Irciniidae (Demospongiae: Dictyoceratida). PLoS ONE, 5(4):e9950.
- Purnomo, H. K., Yusniawati, Y., Putrika, A., Handayani, W., & Yasman. (2017). Seagrass species diversity at various seagrass bed ecosystems in the West Bali National Park area. Prosiding Seminar Nasional Masyarakat Biodiversitas Indonesia, 3(2):236-240.
- Putra, I. N. G., & Putra, I. D. N. N. (2019). Recent invasion of the endemic Banggai cardinalfish, Pterapogon kauderni at the strait of Bali: assessment of the habitat type and population structure. Indonesian Journal of Marine Sciences, 24(1):15-22.
- Reid, B., Le, M., McCord, W., Iverson, J., Georges, A., Bergmann, T., Amto, G., Desalle, R., & Naro-Maciel, E. (2011). Comparing and combining distance-based and character based approaches for barcoding turtles. Molecular Ecology Resources, 11(6):956-967.
- Rhyne, A. L., Tlusty, M. F., Szczebak, J. T., & Holmberg, R. J. (2017). Expanding our understanding of the trade in marine aquarium animals. PeerJ, 5:e2949.
- Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S., & Sánchez-Gracia, S. (2017). DnaSP 6 : DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution, 34(12):3299-3302.
- Saravanan, R., Vijayanand, P., Vagelli, A. A., Murugan, A., Shanker, S., Rajagopal, S., & Balasubramanian, T. (2013). Breeding and rearing of the two striped cardinalfish, Apogon quadrifasciatus (Cuvier, 1828) in captive condition. Animal Reproduction Science, 137(3-4):237-244.
- Sato, M., Nakae, M., & Sasaki, K. (2021). The paedomorphic lateral line system in Pseudamiops and Gymnapogon (Percomorpha, Apogonidae), with morphological and molecular"based phylogenetic considerations. Journal of Morphology, 282(5):652-678.
- Silvestro, D., & Michalak, I. (2012). raxmlGUI: a graphical front-end for RAxML. Organisms Diversity & Evolution, 12:335-337.
- Stamatakis, A. (2014). RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30(9):1312-1313.
- Strona, G., Lafferty, K. D., Fattorini, S., Beck, P. S. A., Guilhaumon, F., Arrigoni, R., Montano, S., Seveso, D., Galli, P., Planes, S., & Parravicini, V. (2021). Global tropical reef fish richness could decline by around half if corals are lost. Proceedings of the Royal Society B, 288(1953):1-8.
- Sultana, M. A., Pandit, D., Barman, S. K., Tikadar, K. K., Tasnim, N., Fagun, I. A., Hussain, M. A., & Kunda, M. (2022). A review of fish diversity, decline drivers, and management of the Tanguar Haor ecosystem: A globally recognized Ramsar site in Bangladesh. Heliyon, 8(11):e11875.
- Tambunan, S., Arthana, I. W., & Putra, I. N. G. (2022). Correlation of density of Banggai Cardinalfish (Pterapogon kauderni) with associated biota (Diadema setosum and Fibramia thermalis) in Gilimanuk Bay, Bali. Journal of Marine Research and Technology, 5(1):16-20.
- Thacker, C. E., & Roje, D. M. (2009). Phylogeny of cardinalfishes (Teleostei: Gobiiformes: Apogonidae) and the evolution of visceral bioluminescence. Molecular Phylogenetics and Evolution, 52(3):735-745.
- Thompson, J., Higgins, D., & Gibson, T. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22(22):4673-4680.
- Thu, P. T., Huang, W. C., Chou, T. K., Van Quan, N., Van Chien, P., Li, F., Shao, K. T., & Liao, T. Y. (2019). DNA barcoding of coastal ray-finned fishes in Vietnam. PLoS ONE, 14(9):1-13.
- Truszkowski, J., & Goldman, N. (2016). Maximum Likelihood phylogenetic inference is consistent on multiple sequence alignments, with or without gaps. Systematic Biology, 65(2):328-333.
- Vagelli, A. A. (2011). The Banggai Cardinalfish: Natural history, conservation, and culture of Pterapogon kauderni. New Jersey: John Wiley & Sons, Ltd.
- Walsh, P. S., Metzger, D. A., & Higuchi, R. (1991). Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques, 54(3):506-513.
- Wang, L., Wu, Z., Liu, M., Liu, W., Zhao, W., Liu, H., & You, F. (2018). DNA barcoding of marine fish species from Rongcheng Bay, China. PeerJ, 6:1-19.
- Ward, R. D., Zemlak, T. S., Innes, B. H., Last, P. R., & Hebert, P. D. N. (2005). DNA barcoding Australia's fish species. Philosophical Transactions of the Royal Society B: Biological Sciences, 360(1462):1847-1857.
References
Abecia, J. E. D., Luiz, O. J., & King, A. J. (2018). Intraspecific morphological and reproductive trait variation in mouth almighty Glossamia aprion (Apogonidae) across different flow environments. Journal of Fish Biology, 93(5):961-971.
Afiati, N., Subagiyo, Handayani, C. R., Hartati, R., & Kholilah, N. (2022). DNA barcoding on Indian Ocean squid, Uroteuthis duvaucelii (D'Orbigny, 1835) (Cephalopoda: Loliginidae) from the Java Sea, Indonesia. Jurnal Ilmiah Perikanan dan Kelautan, 14(2):231-245.
Alfonsi, E., Méheust, E., Fuchs, S., Carpentier, F. G., Quillivic, Y., Viricel, A., Hassani, S., & Jung, J. L. (2013). The use of DNA barcoding to monitor the marine mammal biodiversity along the French Atlantic coast. Zookeys, 365:5-24.
Allen, G. R., & Erdmann, M. V. (2012a). Reef fishes of Bali, Indonesia. In P. L. K. Mustika, I. M. J. Ratha, and S. Purwanto (Ed.), The 2011 Bali marine rapid assessment (Second English edition). (pp. 15-68). Bali: Conservation International.
Allen, G. R., & Erdmann, M. V. (2012b). Reef Fishes of the East Indies. Volume I-III. Virginia: Conservation International Foundation.
Alvarenga, M., Solé-Cava, A. M., & Henning, F. (2021). What's in a name? Phylogenetic species identification reveals extensive trade of endangered guitarfishes and sharks. Biological Conservation, 257:109-119.
Arbi, U. Y., Suharti, S. R., Huwae, R., Rizqi, M. P., & Suratno. (2019). Populasi ikan endemik Capungan Banggai (Pterapogon kauderni) di habitat introduksi di teluk Gilimanuk, Bali. Paper presented at the Seminar Nasional Tahunan XVI Hasil Penelitian Perikanan dan Kelautan, Universitas Gadjah Mada, Indonesia.
Arroyave, J., & Stiassny, M. L. J. (2014). DNA barcoding reveals novel insights into pterygophagy and prey selection in distichodontid fishes (Characiformes: Distichodontidae). Ecology and Evolution, 4(23):4534-4542.
Barber, P. H., Erdmann, M. V, & Palumbi, S. R. (2006). Comparative phylogeography of three codistributed stomatopods: origins and timing of regional lineage diversification in the Coral Triangle. Evolution, 60(9):1825-1839.
Bingpeng, X., Heshan, L., Zhilan, Z., Chunguang, W., Yanguo, W., & Jianjun, W. (2018). DNA barcoding for identification of fish species in the Taiwan Strait. PLoS ONE, 13(6):e0198109.
Cappenberg, H. A. W., Aznam, A., & Aswandy, I. (2006). Komunitas moluska di perairan Teluk Gilimanuk, Bali Barat. Oseanologi dan Liminologi di Indonesia, 40:53-64.
Cinner, J. E., & Mcclanahan, T. R. (2006). Socioeconomic factors that lead to overfishing in small-scale coral reef fisheries of Papua New Guinea. Environmental Conservation, 33(1):73-80.
Delrieu-Trottin, E., Durand, J. D., Limmon, G., Sukmono, T., Kadarusman, Sugeha, H. Y., Chen, W. J., Busson, F., Borsa, P., Dahruddin, H., Sauri, S., Fitriana, Y., Zein, M. S. A., Hocdé, R., Pouyaud, L., Keith, P., Wowor, D., Steinke, D., Hanner, R., & Hubert, N. (2020). Biodiversity inventory of the grey mullets (Actinopterygii: Mugilidae) of the Indo-Australian Archipelago through the iterative use of DNA-based species delimitation and specimen assignment methods. Evolutionary Applications, 13(6):1451-1467.
Fadli, N., Nor, S. A. M., Othman, A. S., Sofyan, H., & Muchlisin, Z. A. (2020). DNA barcoding of commercially important reef fishes in Weh Island, Aceh, Indonesia. PeerJ, 8(2012):1-25.
Falah, I. N., Adharini, R. I., & Ratnawati, S. E. (2023). Molecular identification of elvers (Anguilla spp.) from river estuaries in Central Java, Indonesia using DNA barcoding based on mtDNA CO1 sequences. Jurnal Ilmiah Perikanan dan Kelautan, 15(1):121–130.
Fraser, T. H. (2006). A new species of cardinalfish (Perciformes: Apogonidae: Apogon) from New Caledonia, with comments and a key to related species. Proceedings of the Biological Society of Washington, 119(1):137-142.
Fraser, T. H., Bogorodsky, S. V., Mal, A. O., & Alpermann, T. J. (2021). Review of the cardinalfishes of the genus Cercamia (Percomorpha: Apogonidae) of the Red Sea and Indian Ocean with descriptions of three new species. Zootaxa, 5039(3):363-394.
Fraser, T. H., Randall, J. E., & Allen, G. R. (2002). Clarification of the cardinalfishes (Apogonidae) previously confused with Apogon moluccensis valenciennes, with a description of a related new species. The Raffles Bulletin of Zoology, 50(1):175-184.
Gardiner, N. M., & Jones, G. P. (2005). Habitat specialisation and overlap in a guild of coral reef cardinalfishes (Apogonidae). Marine Ecology Progress Series, 305:163-175.
Gon, O., & Allen, G. R. (2012). Revision of the Indo-Pacific cardinalfish genus Siphamia (Perciformes: Apogonidae). Zootaxa, 3294(1):1-84.
Keyse, J., Treml, E. A., Huelsken, T., Barber, P. H., Deboer, T., Kochzius, M., Nuryanto, A., Gardner, J. P. A., Liu, L. L., Penny, S., & Riginos, C. (2018). Historical divergences associated with intermittent land bridges overshadow isolation by larval dispersal in co-distributed species of Tridacna giant clams. Journal of Biogeography, 45(4):848-858.
Kimura, M. (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 16:111-120.
Kumar, S., Stecher, G., Li, M., Knyaz, C., & Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Molecular Biology and Evolution, 35(6):1547-1549.
Landi, M., Dimech, M., Arculeo, M., Biondo, G., Martins, R., Carneiro, M., Carvalho, G. R., Lo Brutto, S., & Costa, F. O. (2014). DNA barcoding for species assignment: The case of Mediterranean marine fishes. PLoS ONE, 9(9):e106135.
Lazuardi, M. E., Sudiarta, I. K., Ratha, I. M. J., Ampou, E. E., Nugroho, S. C., & Mustika, P. L. (2012). The status of coral reefs in Bali. In P. L. K. Mustika, I. M. J. Ratha, and S. Purwanto (Ed.), The 2011 Bali marine rapid assessment (Second English edition). (pp. 69-77). Bali: Conservation International.
Lea, R. N., Fraser, T. H., Baldwin, C. C., & Craig, M. T. (2022). Five valid species of Cardinalfishes of the Genus Apogon (Apogonidae) in the Eastern Pacific Ocean, with a redescription of A. atricaudus and notes on the distribution of A. atricaudus and A. atradorsatus. Ichthyology & Herpetology, 110(1):106-114.
Lessios, H. A., Kane, J., & Robertson, D. R. (2003). Phylogeography of the pantropical sea urchin Tripneustes: contrasting patterns of population structure between oceans. Evolution, 57(9):2026.
Letunic, I., & Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Research, 49(W1):W293-W296.
Limmon, G., Delrieu-Trottin, E., Patikawa, J., Rijoly, F., Dahruddin, H., Busson, F., Steinke, D., & Hubert, N. (2020). Assessing species diversity of coral triangle artisanal fisheries: A DNA barcode reference library for the shore fishes retailed at Ambon harbor (Indonesia). Ecology and Evolution, 10(7):3356-3366.
Luehrmann, M., Cortesi, F., Cheney, K. L., de Busserolles, F., & Marshall, N. J. (2020). Microhabitat partitioning correlates with opsin gene expression in coral reef cardinalfishes (Apogonidae). Functional Ecology, 34(5):1041-1052.
Ma'ruf, M. S., Arthana, I. W., & Ernawati, N. M. (2022). Composition of types and conditions of mangrove in Gilimanuk Bay, West Bali National Park. Ecotrophic, 16(2):153-173.
Maas, D. L., Capriati, A., Ahmad, A., Erdmann, M. V., Lamers, M., de Leeuw, C. A., Prins, L., Purwanto, Putri, A. P., Tapilatu, R. F., & Becking, L. E. (2020). Recognizing peripheral ecosystems in marine protected areas: A case study of golden jellyfish lakes in Raja Ampat, Indonesia. Marine Pollution Bulletin, 151:110700.
Mabuchi, K., Fraser, T. H., Song, H., Azuma, Y., & Nishida, M. (2014). Revision of the systematics of the cardinalfishes (Percomorpha: Apogonidae) based on molecular analyses and comparative reevaluation of morphological characters. Zootaxa, 3846(2):151-203.
Mbaba, K. Y., Suryatika, I. B. M., & Adnyana, I. G. A. P. (2019). Analysis of the concentration of Cd metal in Rhizophora.Sp using the AAS method in the Gilimanuk Harbor area, Jembrana, Bali. Kappa Journal, 3:83-88.
Meyers-Muñoz, M. A., van der Velde, G., van der Meij, S. E. T., Stoffels, B. E. M. W., van Alen, T., Tuti, Y., & Hoeksema, B. W. (2016). The phylogenetic position of a new species of Plakobranchus from West Papua, Indonesia (Mollusca, Opisthobranchia, Sacoglossa). Zookeys, 594:73-98.
Nei, M., & Kumar, S. (2000). Molecular evolution and phylogenetics. New York: Oxford University Press.
Pamungkas, P. B. P., Hendrawan, I. G., & Putra, I. N. G. (2021). Characteristics and distribution of stranded waste in the coastal area of West Bali National Park. Journal of Marine Research and Technology, 4(1):9-15.
Permana, G. N., Slamet, B., Permana, B. A., Dewi, A. K., & Mahardika, G. N. (2019). Population genetic structure of spiny lobsters, Panulirus homarus and Panulirus ornatus, in the Indian Ocean , Coral Triangle, and South China Sea. Indonesian Aquaculture Journal, 14(1):7-14.
Pettaloloa, D., Watiniasih, N., & Sari, A. (2022). The diversity of water bird species in the Karang Sewu mangrove forest, Gilimanuk, Bali. Current Trends in Aquatic Science, 3:45-51.
Pöppe, J., Sutcliffe, P., Hooper, J. N. A., Wörheide, G., & Erpenbeck, D. (2010). COI Barcoding reveals new clades and radiation patterns of Indo-Pacific sponges of the family Irciniidae (Demospongiae: Dictyoceratida). PLoS ONE, 5(4):e9950.
Purnomo, H. K., Yusniawati, Y., Putrika, A., Handayani, W., & Yasman. (2017). Seagrass species diversity at various seagrass bed ecosystems in the West Bali National Park area. Prosiding Seminar Nasional Masyarakat Biodiversitas Indonesia, 3(2):236-240.
Putra, I. N. G., & Putra, I. D. N. N. (2019). Recent invasion of the endemic Banggai cardinalfish, Pterapogon kauderni at the strait of Bali: assessment of the habitat type and population structure. Indonesian Journal of Marine Sciences, 24(1):15-22.
Reid, B., Le, M., McCord, W., Iverson, J., Georges, A., Bergmann, T., Amto, G., Desalle, R., & Naro-Maciel, E. (2011). Comparing and combining distance-based and character based approaches for barcoding turtles. Molecular Ecology Resources, 11(6):956-967.
Rhyne, A. L., Tlusty, M. F., Szczebak, J. T., & Holmberg, R. J. (2017). Expanding our understanding of the trade in marine aquarium animals. PeerJ, 5:e2949.
Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S., & Sánchez-Gracia, S. (2017). DnaSP 6 : DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution, 34(12):3299-3302.
Saravanan, R., Vijayanand, P., Vagelli, A. A., Murugan, A., Shanker, S., Rajagopal, S., & Balasubramanian, T. (2013). Breeding and rearing of the two striped cardinalfish, Apogon quadrifasciatus (Cuvier, 1828) in captive condition. Animal Reproduction Science, 137(3-4):237-244.
Sato, M., Nakae, M., & Sasaki, K. (2021). The paedomorphic lateral line system in Pseudamiops and Gymnapogon (Percomorpha, Apogonidae), with morphological and molecular"based phylogenetic considerations. Journal of Morphology, 282(5):652-678.
Silvestro, D., & Michalak, I. (2012). raxmlGUI: a graphical front-end for RAxML. Organisms Diversity & Evolution, 12:335-337.
Stamatakis, A. (2014). RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30(9):1312-1313.
Strona, G., Lafferty, K. D., Fattorini, S., Beck, P. S. A., Guilhaumon, F., Arrigoni, R., Montano, S., Seveso, D., Galli, P., Planes, S., & Parravicini, V. (2021). Global tropical reef fish richness could decline by around half if corals are lost. Proceedings of the Royal Society B, 288(1953):1-8.
Sultana, M. A., Pandit, D., Barman, S. K., Tikadar, K. K., Tasnim, N., Fagun, I. A., Hussain, M. A., & Kunda, M. (2022). A review of fish diversity, decline drivers, and management of the Tanguar Haor ecosystem: A globally recognized Ramsar site in Bangladesh. Heliyon, 8(11):e11875.
Tambunan, S., Arthana, I. W., & Putra, I. N. G. (2022). Correlation of density of Banggai Cardinalfish (Pterapogon kauderni) with associated biota (Diadema setosum and Fibramia thermalis) in Gilimanuk Bay, Bali. Journal of Marine Research and Technology, 5(1):16-20.
Thacker, C. E., & Roje, D. M. (2009). Phylogeny of cardinalfishes (Teleostei: Gobiiformes: Apogonidae) and the evolution of visceral bioluminescence. Molecular Phylogenetics and Evolution, 52(3):735-745.
Thompson, J., Higgins, D., & Gibson, T. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22(22):4673-4680.
Thu, P. T., Huang, W. C., Chou, T. K., Van Quan, N., Van Chien, P., Li, F., Shao, K. T., & Liao, T. Y. (2019). DNA barcoding of coastal ray-finned fishes in Vietnam. PLoS ONE, 14(9):1-13.
Truszkowski, J., & Goldman, N. (2016). Maximum Likelihood phylogenetic inference is consistent on multiple sequence alignments, with or without gaps. Systematic Biology, 65(2):328-333.
Vagelli, A. A. (2011). The Banggai Cardinalfish: Natural history, conservation, and culture of Pterapogon kauderni. New Jersey: John Wiley & Sons, Ltd.
Walsh, P. S., Metzger, D. A., & Higuchi, R. (1991). Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques, 54(3):506-513.
Wang, L., Wu, Z., Liu, M., Liu, W., Zhao, W., Liu, H., & You, F. (2018). DNA barcoding of marine fish species from Rongcheng Bay, China. PeerJ, 6:1-19.
Ward, R. D., Zemlak, T. S., Innes, B. H., Last, P. R., & Hebert, P. D. N. (2005). DNA barcoding Australia's fish species. Philosophical Transactions of the Royal Society B: Biological Sciences, 360(1462):1847-1857.