Date Log
Copyright (c) 2024 Jurnal Ilmiah Perikanan dan Kelautan

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
1. Copyright of the article is transferred to the journal, by the knowledge of the author, whilst the moral right of the publication belongs to the author.
2. The legal formal aspect of journal publication accessibility refers to Creative Commons Atribusi-Non Commercial-Share alike (CC BY-NC-SA), (https://creativecommons.org/licenses/by-nc-sa/4.0/)
3. The articles published in the journal are open access and can be used for non-commercial purposes. Other than the aims mentioned above, the editorial board is not responsible for copyright violation
The manuscript authentic and copyright statement submission can be downloaded ON THIS FORM.
Genetic Diversity and Population Structure of Amphidromous Goby (Stiphodon semoni) in Western Part of Southern Java Waters
Corresponding Author(s) : Ahmad Romdon
Jurnal Ilmiah Perikanan dan Kelautan, Vol. 16 No. 2 (2024): JURNAL ILMIAH PERIKANAN DAN KELAUTAN
Abstract
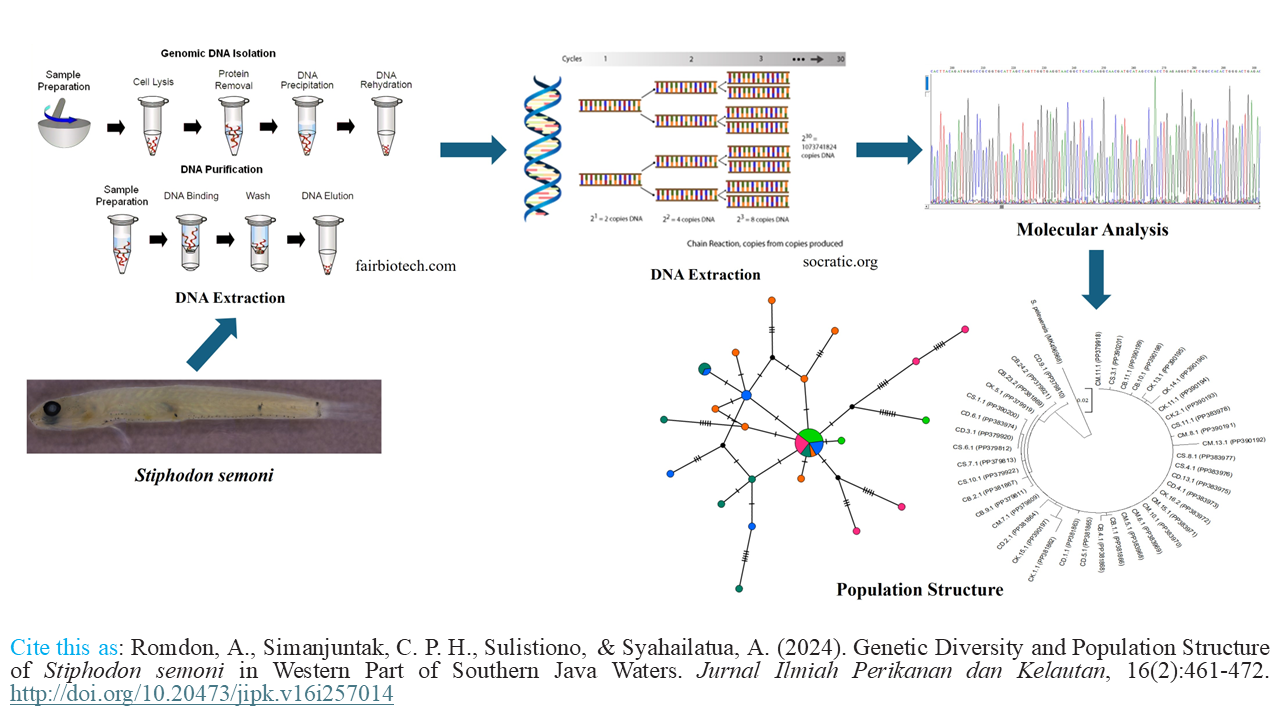
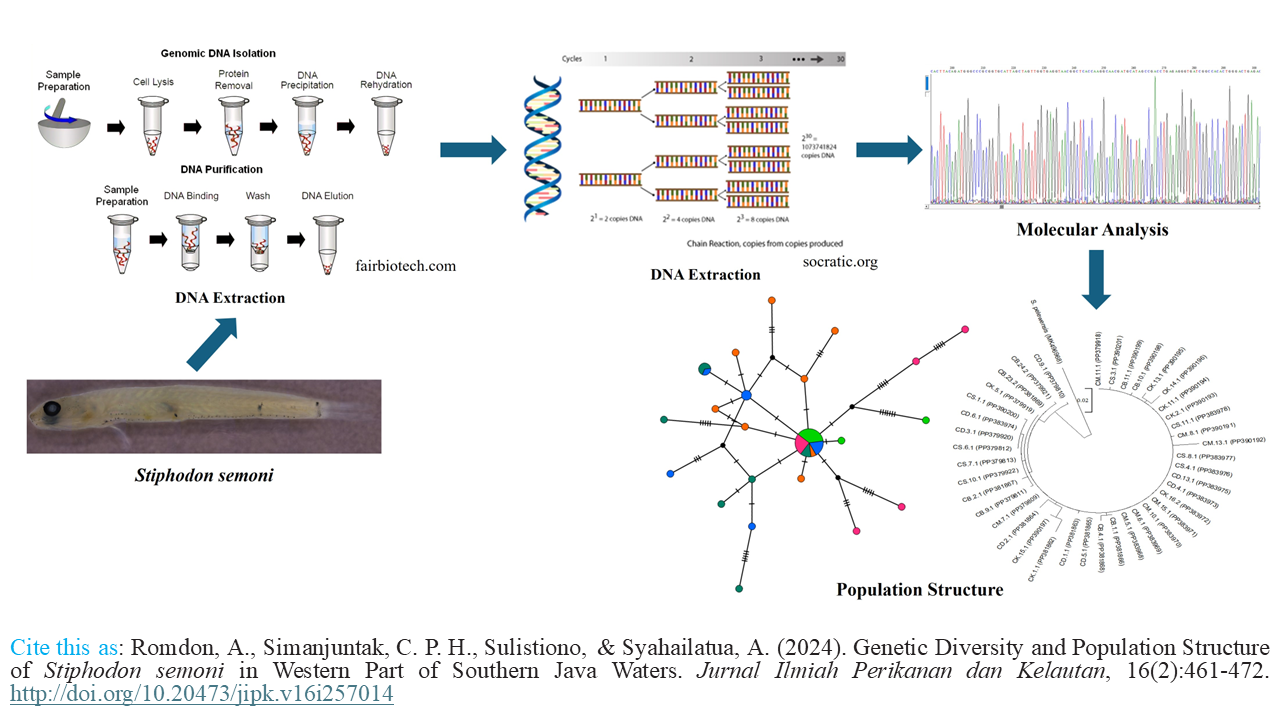
Graphical Abstract
Research Highlight
- The genetic diversity of Stiphodon semoni is high.
- All populations have high haplotype diversity except for Cimaja.
- No genetic structure is observed within the populations of semoni.
- The population of semoni has undergone demographic expansion.
- Cibareno River has the potential to be a protected area.
Abstract
Fishing activities negatively impact fish populations, potentially causing a decline in fish stocks. Nevertheless, ensuring diversity and connectivity among populations can mitigate these adverse effects. To evaluate the connectivity of river mouths in the western part of Southern Java waters, we sequenced forty Stiphodon semoni individuals from five populations using mitochondrial cytochrome oxidase subunit 1 as molecular markers. The study revealed that S. semoni populations showed high diversity (0.821), with the population in Cimaja displaying the lowest diversity (0.464). Furthermore, the result of the analysis of molecular variance was a Fst value of 0.0630 with a p-value of 0.22. These results along with the result of the haplotype network indicated no significant genetic differences among these populations. This implies that the river mouths in the western part of Southern Java waters are interconnected. The distribution of mismatches showed a single peak, indicating that the populations have undergone demographic expansion. This information could be valuable for the conservation and management of S. semoni in the western part of the Southern Java waters.
Keywords
Download Citation
Endnote/Zotero/Mendeley (RIS)BibTeX
- Abdulmalik-Labe, O. P., Eduardo, A. J. L., & Quilang, J. P. (2023). Genetic diversity and divergence among native and translocated populations of the golden flathead goby Glossogobius aureus (Gobiiformes: Gobiidae) in Philippine lakes. PLoS ONE, 18(12):1-25.
- Afiati, N., Subagiyo, Handayani, C. R., Hartati, R., & Kholilah, N.. (2022). DNA barcoding on Indian Ocean Squid, Uroteuthis duvaucelii (D’Orbigny, 1835) (Cephalopoda: Loliginidae) from the Java Sea, Indonesia. Jurnal Ilmiah Perikanan dan Kelautan, 14(2):231-245.
- Alexandrino, P. B., Faria, R., Castro, F., Le Corre, M., Sabatié, R., Baglinière, J. L., & Weiss, S. (2006). Interspecifc diferentiation and intraspecifc substructure in two closely related clupeids with extensive hybridization, Alosa alosa and Alosa fallax. Journal of Fish Biology, 69(1):242-259.
- Amaliah, S. W., Affandi, R., Simanjuntak, C. P. H., Baihaqi, F., Annida, S. B., Prabowo, T., & Romdon, A. (2023). Diet Composition and Feeding Strategy Of Larvae and Juveniles Of Green Riffle Goby, Stiphodon elegans in Cimaja Estuary, Indonesia. IOP Conference Series: Earth Environmental Science, 1033(012009):1-11.
- Baihaqi, F., Simanjuntak, C. P. H., Sulistiono, Prabowo, T., Annida, S. B., Ervinia, A., & Budiman M. S. (2022). Distribution, abundance, and species composition of fish larvae and juveniles of Gobiidae in the Cimaja estuary, Palabuhanratu, Indonesia. IOP Conference Series: Earth Environmental Science, 1033(012004):1-12.
- Bandelt, H. J., Forster, P., & Rohl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16(1):37-48.
- Bashalkhanov, S., Pandey, M., & Rajora, O. P. (2009). A simple method for estimating genetic diversity in large populations from finite sample sizes. BMC Genetics, 10(84):1-10
- Berkström, C., Wennerström, L., & Bergström, U. (2022). Ecological connectivity of the marine protected area network in the Baltic Sea, Kattegat and Skagerrak: Current knowledge and management needs. Ambio, 51(1):1485-503.
- Cheng, F., Wang, Q., Delser, P. M., & Li, C. (2019). Multiple freshwater invasions of the tapertail anchovy (Clupeiformes: Engraulidae) of the Yangtze River. Ecology and Evolution, 9(21): 12202-12215.
- Eastwood, E. K., Lopez, E. H., & Drew, J. A. (2016). Population connectivity measures of fishery-targeted coral reef species to inform marine reserve network design in Fiji. Scientific Reports, 6(1):1-10.
- Engman, A. C., Hogue, G. M., Starnes, W. C., Raley, M. E., & Kwak, T. J. (2019). Puerto Rico Sicydium goby diversity: species-specific insights on population structures and distributions. Neotropical Biodiversity, 5(1):22-29. https://doi.org/10.1080/23766808.2019.1606669.
- Excoffier, L., Smouse, P. E., & Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics, 131(1):479-491.
- Faria, R., Weiss, S., & Alexandrino, P. B. (2012). Comparative phylogeography and demographic history of European shads (Alosa alosa and A. fallax) inferred from mitochondrial DNA. BMC Evolutonary Biology, 12(194):1-20.
- Galtier, N., Nabholz, B., Glémin, S., & Hurst, G. D. D. (2009). Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Molecular Ecology, 18(22):4541-4550.
- Garcia, E., Wright, D., Gatins, R., Roberts, M. B., Pinheiro, H. T., Salas, E., Chen, J-Y., Winnikoff, J. R., & Bernardi, G. (2021). Haplotype network branch diversity, a new metric combining genetic and topological diversity to compare the complexity of haplotype networks. PLoS ONE, 16(6): 1-15.
- Gorbachev, V. V. (2012). Effect of random sample size on the accuracy of nucleotide diversity estimation. Russ. J. Genet, 48(7):747-751. https://doi.org/10.1134/S1022795412050079.
- Hall, T. A. (1999). BIOEDIT: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/ NT. Nucleic Acids Symposium Series, 41:95-98.
- Hall, A. E., Vitale, L., & Kingsford, M. J. (2019). Planktonic larval duration, early growth, and the influence of dietary input on the otolith microstructure of Scolopsis bilineatus (Nemipteridae). Environmental Biology of Fishes, 102(1):541-552.
- Hidayani, A. A., Tasakka, A. C. M. A. R., Umar, W., Alam, M. J., Neogi, A. K., & Andriyono, S. (2022). Low genetic diversity study on leopard coral grouper Plectropomus leopardus (Perciformes: Serranidae) from Sulawesi, Indonesia. Jurnal Ilmiah Perikanan dan Kelautan, 14(2):349-359. http://doi.org/10.20473/jipk.v14i2.32815
- Hoareau, T. B., Lecomte-Finiger, R., Grondin, H. P., Conand, C., & Berrebi, P. (2007). Oceanic larval life of La Reunion ’bichiques’, amphidromous gobiid post-larvae. Marine Ecology Progress Series, 333:303-308.
- Hohenlohe, P. A. (2004). Limits to gene flow in marine animals with planktonic larvae: Models of Littorina species around Point Conception, California. Biological Journal of the Linnean Society, 82(2):169-87.
- Hudson, R. R., Slatkin, M., & Maddison, W. P. (1992). Estimation of levels of gene flow from DNA sequence data. Genetics, 132(1):583-589. doi: 10.1093/genetics/132.2.583.
- Hughes, A. R., Inouye, B. D., Johnson, M. T. J., Underwood, N., & Vellend, M. (2008). Ecological consequences of genetic diversity. Ecology Letters, 11(6):609-623
- Jukes, T. H., & Cantor, C. R. (1969). Evolution of Protein Molecules. In: Munro, H.N., Ed., Mammalian Protein Metabolism. New York: Academic Press. 21-132.
- Lohmann, K. J., & Lohmann, C. M. F. (2019). There and back again: natal homing by magnetic navigation in sea turtles and salmon. Journal of Experimental Biology, 222(Suppl 1): jeb184077.
- Kitada, S. (2022). Long-term translocation explains population genetic structure of a recreationally fished iconic species in Japan: Combining current knowledge with reanalysis. Aquaculture, Fish and Fisheries, 2(1):130-145.
- Klerks, P. L., Athrey, G. N., & Leberg, P. L. (2019). Response to selection for increased heat tolerance in a small fish species, with the response decreased by a population bottleneck. Frontiers in Ecology and Evolution, 7(270):1-20.
- Leis, J. M., & Carson-Ewart, B. M. (2004). The larvae of Indo-Pacific coastal fishes: An identification guide to marine fish larvae. 2nd Edition. Leiden: Brill.
- Leigh, J. W., & Bryant, D. (2015). Popart: Full-feature software for haplotype network construction. Methods in Ecology and Evolution, 6(9):1110-1116.
- Liao, T-Y., Lu, P-L., Yu, Y-H., Huang, W-C., Shiao, J-C., Lin, H-D., Jhuang, W-C., Chou, T-K., & Li, F. (2021) Amphidromous but endemic: Population connectivity of Rhinogobius gigas (Teleostei: Gobioidei). PLoS ONE, 16(2):1-14.
- Lisi, P. J., Hogan, J. D., Holt, G., Moody, K. N., Wren, J. L. K., Kobayashi, D. R., Blum, M. J., & McIntyre, P. B. (2022). Stream and ocean hydrodynamics mediate partial migration strategies in an amphidromous Hawaiian goby. Ecology, 103(11):1-13.
- Liu, S., Xue, Q., Xu, H., & Lin, Z. (2021). Identification of main oyster species and comparison of their genetic diversity in Zhejiang Coast, south of Yangtze River estuary. Frontiers in Marine Science, 8(662515):1-13.
- Lord, C., Brun, C., Hautecoeur, M., & Keith, P. (2010). Insights on endemism: Comparison of the duration of the marine larval phase estimated by otolith microstructural analysis of three amphidromous Sicyopterus species (Gobioidei: Sicydiinae) from Vanuatu and New Caledonia. Ecology of Freshwater Fish, 19(1):26-38.
- Lord, C., Lorion, J., Dettai, A., Watanabe, S., Tsukamoto, K., Cruaud, C., & Keith, P. (2012). From endemism to widespread distribution: Phylogeography of three amphidromous Sicyopterus species (Teleostei: Gobioidei: Sicydiinae). Marine Ecology Progress Series, 455(1):269-285.
- Lord, C., Maeda, K., Keith, P., & Watanabe, S. (2015). Population structure of the asian amphidromous Sicydiinae goby, Stiphodon percnopterygionus, inferred from mitochondrial COI sequences, with comments on larval dispersal in the Northwest Pacific Ocean. Vie Milieu, 65(2):63-71.
- Madduppa, H., Martaulina, R., Zairion, Z., Renjani, R. M., Kawaroe, M., Anggraini, N. P., Subhan B., Verawati I., & Sani, L. M. I. (2021). Genetic population subdivision of the blue swimming crab (Portunus pelagicus) across Indonesia inferred from mitochondrial DNA: Implication to sustainable fishery. PLoS ONE, 16(2):1-14.
- Maeda, K., & Tachihara, K. (2005). Recruitment of amphidromous sleepers Eleotris acanthopoma, Eleotris melanosoma, and Eleotris fusca into the Teima River, Okinawa Island. Ichthyologcal Research, 52(4):325-335.
- McDowall, R. M. (2001). Anadromy and homing: two life-history traits with adaptive synergies in salmonid fishes? Fish Fisheries, 2(1):78-85.
- McDowall, R. M. (2007). On amphidromy, a distinct form of diadromy in aquatic organisms. Fish Fisheries, 8(1):1-13.
- Modeel, S., Joshi, B. D., Yadav, S., Bharti, M., & Negi, R. K. (2023). Mitochondrial DNA reveals shallow population genetic structure in economically important Cyprinid fish Labeo rohita (Hamilton, 1822) from South and Southeast Asia. Molecular Biology Reports, 50(6):4759-4767.
- O'Dwyer, J. E., Murphy, N., Tonkin, Z., Lyon, J., Koster, W., Dawson, D., Amtstaetter, F., & Harrisson, K. A. (2021). An investigation of genetic connectivity shines a light on the relative roles of isolation by distance and oceanic currents in three diadromous fish species. Marine and Freshwater Research, 72(10):1457-1473.
- Orlova, S. Y., Emelyanova, O. R., Nebesikhina, N. A. Rabazanov, N. I., & Orlov, A. M. (2024). The problems of DNA-barcoding the shads of genus Alosa (Alosidae) of the Ponto-Caspian Basin. Journal of Ichthyology, 64(3):510-520.
- Perrier, C., Ferchaud, A-L., Sirois, P., Thibault, I., & Bernatchez, l. (2017). Do genetic drift and accumulation of deleterious mutations preclude adaptation? Empirical investigation using RADseq in a northern lacustrine fish. Molecular Ecology, 26(22):6317-6335.
- Peery, M. Z., Kirby, R., Reid, B. N., Stoelting, R., Doucet-Bëer, E., Robinson, S., Vásquez-Carrillo, C., Pauli, J. N., & Palsbøll, P. J. (2012). Reliability of genetic bottleneck tests for detecting recent population declines. Molecular Ecology, 21(14):3403-3418.
- Pinacho-Pinacho, C., D., Guzmán-Valdivieso, I., Calixto-Rojas, M., García-Vásquez, A., & Rubio-Godoy, M. (2023). Morphological and molecular characterization of three new species of Gyrodactylus (Monogenea: Gyrodactylidae) infecting Sicydium salvini (Teleostei: Gobiidae) in Mexican rivers draining into the Pacific Ocean. Parasitology Internasional, 93(102712):1-12.
- Prabowo, T., Simanjuntak, C. P. H., Affandi, R., Baihaqi, F., Annida, S. B., Ervinia, A., & Budiman, M. S. (2022). Temporal variation in abundance of bully sleepers (Pisces: Eleotridae) larvae and juveniles in Cisolok Estuary, Palabuhanratu Bay, Indonesia. IOP Conference Series: Earth Environmental Science, 1033(012003):1-9.
- Ramírez-Soriano, A., Ramos-Onsins, S. E., Rozas, J., Calafell, F., & Navarro, A. (2008). Statistical power analysis of neutrality tests under demographic expansions, contractions and bottlenecks with recombination. Genetics, 179(1):555-567.
- Ramos-Onsins, S. E., & Rozas, J. (2006). Statistical properties of new neutrality tests against population growth. Molecular Biology and Evolution, 23(8):2092-2100.
- Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., & Sánchez-Gracia, A. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution, 34(12):3299-3302.
- Sabatino, S. J., Faria, R., & Alexandrino, P. B. (2022). Genetic structure, diversity, and connectivity in anadromous and freshwater Alosa alosa and A. fallax. Marine Biology, 169(1):1-14.
- Sahami, F. M., Kepel, R. C., Olii, A. H., Pratasik, S. B., Lasabuda, R., Wantasen, A., & Habibie, S. A. (2020). Morphometric and genetic variations of species composers of nike fish assemblages in Gorontalo Bay Waters, Indonesia. Biodiversitas, 21(10):4571-4581.
- Salmenkova, E., A. (2017). Mechanisms of homing in salmonids. Biology Bulletin Reviews, 7(4):287-298.
- Sánchez-Montes, G., Ariño, A. H., Vizmanos, J. L., Wang, J., & Martínez-Solano, Í. (2017). Effects of sample size and full sibs on genetic diversity characterization: A case study of three syntopic iberian pond-breeding amphibians. Journal of Heredity, 108(5):535-543.
- Selbie, D. T., Lewis, B. A., Smol, J. P., & Finney, B. P. (2011). Long‐term population dynamics of the endangered snake river sockeye salmon: Evidence of past influences on stock decline and impediments to recovery. Transactions of the American Fisheries Society, 136(3):800-821.
- Simanjuntak, C. P. H., Baihaqi, F., Prabowo, T., Bilqis, A. S., Sulistiono, & Ervinia, A. (2021). Recruitment patterns of freshwater amphidromous fishes (Pisces: Gobiidae, Eleotridae) to the Cimaja estuary, Palabuhanratu Bay. [in Indonesian] Jurnal Iktiologi Indonesia, 21(3):321-337.
- Taillebois, L., Castelin, M., Ovenden, J. R., Bonillo, C., & Keith, P. (2013). Contrasting genetic structure among populations of two amphidromous fish species (Sicydiinae) in the Central West Pacific. PLoS ONE, 8(10):1-13.
- Tamura, K., Stecher, G., & Kumar, S. (2021). MEGA11: Molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution, 38(7):3022-3027.
- Walter, R., Hogan, J., Blum, M., Gagne, R., Hain, E., Gilliam, J., & McIntyre, P. (2012). Climate change and conservation of endemic amphidromous fishes in Hawaiian streams. Endangered Species Research, 16:261-272.
- Wang, M., Zhang, X., Yang, T., Han, Z., Yanagimoto, T., & Gao, T. (2008). Genetic diversity in the mtDNA control region and population structure in the Sardinella zunasi Bleeker. African Journal of Biotechnology, 7(24):4384-4392.
- Watanabe, S., Iida, M., Kimura, Y., Feunteun, E., & Tsukamoto, K. (2006). Genetic diversity of Sicyopterus japonicus as revealed by mitochondrial DNA sequencing. Coastal Marine Science, 30(2):473-479.
- Xuan, Z., Jiang, T., Liu, H., Chen, X., & Yang, J. (2021). Mitochondrial DNA and microsatellite analyses reveal strong genetic differentiation between two types of estuarine tapertail anchovies (Coilia) in Yangtze River Basin, China. Hydrobiologia, 848:1409-1431.
- Zhao, F., Liu, Y., Wang, Z., Lu, J., Cao, L., & Zeng, C. (2023). Genetic diversity and connectivity of Ocypode ceratophthalmus in the East and South China Seas and its implications for conservation. Biology, 12(437):1-13.
References
Abdulmalik-Labe, O. P., Eduardo, A. J. L., & Quilang, J. P. (2023). Genetic diversity and divergence among native and translocated populations of the golden flathead goby Glossogobius aureus (Gobiiformes: Gobiidae) in Philippine lakes. PLoS ONE, 18(12):1-25.
Afiati, N., Subagiyo, Handayani, C. R., Hartati, R., & Kholilah, N.. (2022). DNA barcoding on Indian Ocean Squid, Uroteuthis duvaucelii (D’Orbigny, 1835) (Cephalopoda: Loliginidae) from the Java Sea, Indonesia. Jurnal Ilmiah Perikanan dan Kelautan, 14(2):231-245.
Alexandrino, P. B., Faria, R., Castro, F., Le Corre, M., Sabatié, R., Baglinière, J. L., & Weiss, S. (2006). Interspecifc diferentiation and intraspecifc substructure in two closely related clupeids with extensive hybridization, Alosa alosa and Alosa fallax. Journal of Fish Biology, 69(1):242-259.
Amaliah, S. W., Affandi, R., Simanjuntak, C. P. H., Baihaqi, F., Annida, S. B., Prabowo, T., & Romdon, A. (2023). Diet Composition and Feeding Strategy Of Larvae and Juveniles Of Green Riffle Goby, Stiphodon elegans in Cimaja Estuary, Indonesia. IOP Conference Series: Earth Environmental Science, 1033(012009):1-11.
Baihaqi, F., Simanjuntak, C. P. H., Sulistiono, Prabowo, T., Annida, S. B., Ervinia, A., & Budiman M. S. (2022). Distribution, abundance, and species composition of fish larvae and juveniles of Gobiidae in the Cimaja estuary, Palabuhanratu, Indonesia. IOP Conference Series: Earth Environmental Science, 1033(012004):1-12.
Bandelt, H. J., Forster, P., & Rohl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16(1):37-48.
Bashalkhanov, S., Pandey, M., & Rajora, O. P. (2009). A simple method for estimating genetic diversity in large populations from finite sample sizes. BMC Genetics, 10(84):1-10
Berkström, C., Wennerström, L., & Bergström, U. (2022). Ecological connectivity of the marine protected area network in the Baltic Sea, Kattegat and Skagerrak: Current knowledge and management needs. Ambio, 51(1):1485-503.
Cheng, F., Wang, Q., Delser, P. M., & Li, C. (2019). Multiple freshwater invasions of the tapertail anchovy (Clupeiformes: Engraulidae) of the Yangtze River. Ecology and Evolution, 9(21): 12202-12215.
Eastwood, E. K., Lopez, E. H., & Drew, J. A. (2016). Population connectivity measures of fishery-targeted coral reef species to inform marine reserve network design in Fiji. Scientific Reports, 6(1):1-10.
Engman, A. C., Hogue, G. M., Starnes, W. C., Raley, M. E., & Kwak, T. J. (2019). Puerto Rico Sicydium goby diversity: species-specific insights on population structures and distributions. Neotropical Biodiversity, 5(1):22-29. https://doi.org/10.1080/23766808.2019.1606669.
Excoffier, L., Smouse, P. E., & Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics, 131(1):479-491.
Faria, R., Weiss, S., & Alexandrino, P. B. (2012). Comparative phylogeography and demographic history of European shads (Alosa alosa and A. fallax) inferred from mitochondrial DNA. BMC Evolutonary Biology, 12(194):1-20.
Galtier, N., Nabholz, B., Glémin, S., & Hurst, G. D. D. (2009). Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Molecular Ecology, 18(22):4541-4550.
Garcia, E., Wright, D., Gatins, R., Roberts, M. B., Pinheiro, H. T., Salas, E., Chen, J-Y., Winnikoff, J. R., & Bernardi, G. (2021). Haplotype network branch diversity, a new metric combining genetic and topological diversity to compare the complexity of haplotype networks. PLoS ONE, 16(6): 1-15.
Gorbachev, V. V. (2012). Effect of random sample size on the accuracy of nucleotide diversity estimation. Russ. J. Genet, 48(7):747-751. https://doi.org/10.1134/S1022795412050079.
Hall, T. A. (1999). BIOEDIT: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/ NT. Nucleic Acids Symposium Series, 41:95-98.
Hall, A. E., Vitale, L., & Kingsford, M. J. (2019). Planktonic larval duration, early growth, and the influence of dietary input on the otolith microstructure of Scolopsis bilineatus (Nemipteridae). Environmental Biology of Fishes, 102(1):541-552.
Hidayani, A. A., Tasakka, A. C. M. A. R., Umar, W., Alam, M. J., Neogi, A. K., & Andriyono, S. (2022). Low genetic diversity study on leopard coral grouper Plectropomus leopardus (Perciformes: Serranidae) from Sulawesi, Indonesia. Jurnal Ilmiah Perikanan dan Kelautan, 14(2):349-359. http://doi.org/10.20473/jipk.v14i2.32815
Hoareau, T. B., Lecomte-Finiger, R., Grondin, H. P., Conand, C., & Berrebi, P. (2007). Oceanic larval life of La Reunion ’bichiques’, amphidromous gobiid post-larvae. Marine Ecology Progress Series, 333:303-308.
Hohenlohe, P. A. (2004). Limits to gene flow in marine animals with planktonic larvae: Models of Littorina species around Point Conception, California. Biological Journal of the Linnean Society, 82(2):169-87.
Hudson, R. R., Slatkin, M., & Maddison, W. P. (1992). Estimation of levels of gene flow from DNA sequence data. Genetics, 132(1):583-589. doi: 10.1093/genetics/132.2.583.
Hughes, A. R., Inouye, B. D., Johnson, M. T. J., Underwood, N., & Vellend, M. (2008). Ecological consequences of genetic diversity. Ecology Letters, 11(6):609-623
Jukes, T. H., & Cantor, C. R. (1969). Evolution of Protein Molecules. In: Munro, H.N., Ed., Mammalian Protein Metabolism. New York: Academic Press. 21-132.
Lohmann, K. J., & Lohmann, C. M. F. (2019). There and back again: natal homing by magnetic navigation in sea turtles and salmon. Journal of Experimental Biology, 222(Suppl 1): jeb184077.
Kitada, S. (2022). Long-term translocation explains population genetic structure of a recreationally fished iconic species in Japan: Combining current knowledge with reanalysis. Aquaculture, Fish and Fisheries, 2(1):130-145.
Klerks, P. L., Athrey, G. N., & Leberg, P. L. (2019). Response to selection for increased heat tolerance in a small fish species, with the response decreased by a population bottleneck. Frontiers in Ecology and Evolution, 7(270):1-20.
Leis, J. M., & Carson-Ewart, B. M. (2004). The larvae of Indo-Pacific coastal fishes: An identification guide to marine fish larvae. 2nd Edition. Leiden: Brill.
Leigh, J. W., & Bryant, D. (2015). Popart: Full-feature software for haplotype network construction. Methods in Ecology and Evolution, 6(9):1110-1116.
Liao, T-Y., Lu, P-L., Yu, Y-H., Huang, W-C., Shiao, J-C., Lin, H-D., Jhuang, W-C., Chou, T-K., & Li, F. (2021) Amphidromous but endemic: Population connectivity of Rhinogobius gigas (Teleostei: Gobioidei). PLoS ONE, 16(2):1-14.
Lisi, P. J., Hogan, J. D., Holt, G., Moody, K. N., Wren, J. L. K., Kobayashi, D. R., Blum, M. J., & McIntyre, P. B. (2022). Stream and ocean hydrodynamics mediate partial migration strategies in an amphidromous Hawaiian goby. Ecology, 103(11):1-13.
Liu, S., Xue, Q., Xu, H., & Lin, Z. (2021). Identification of main oyster species and comparison of their genetic diversity in Zhejiang Coast, south of Yangtze River estuary. Frontiers in Marine Science, 8(662515):1-13.
Lord, C., Brun, C., Hautecoeur, M., & Keith, P. (2010). Insights on endemism: Comparison of the duration of the marine larval phase estimated by otolith microstructural analysis of three amphidromous Sicyopterus species (Gobioidei: Sicydiinae) from Vanuatu and New Caledonia. Ecology of Freshwater Fish, 19(1):26-38.
Lord, C., Lorion, J., Dettai, A., Watanabe, S., Tsukamoto, K., Cruaud, C., & Keith, P. (2012). From endemism to widespread distribution: Phylogeography of three amphidromous Sicyopterus species (Teleostei: Gobioidei: Sicydiinae). Marine Ecology Progress Series, 455(1):269-285.
Lord, C., Maeda, K., Keith, P., & Watanabe, S. (2015). Population structure of the asian amphidromous Sicydiinae goby, Stiphodon percnopterygionus, inferred from mitochondrial COI sequences, with comments on larval dispersal in the Northwest Pacific Ocean. Vie Milieu, 65(2):63-71.
Madduppa, H., Martaulina, R., Zairion, Z., Renjani, R. M., Kawaroe, M., Anggraini, N. P., Subhan B., Verawati I., & Sani, L. M. I. (2021). Genetic population subdivision of the blue swimming crab (Portunus pelagicus) across Indonesia inferred from mitochondrial DNA: Implication to sustainable fishery. PLoS ONE, 16(2):1-14.
Maeda, K., & Tachihara, K. (2005). Recruitment of amphidromous sleepers Eleotris acanthopoma, Eleotris melanosoma, and Eleotris fusca into the Teima River, Okinawa Island. Ichthyologcal Research, 52(4):325-335.
McDowall, R. M. (2001). Anadromy and homing: two life-history traits with adaptive synergies in salmonid fishes? Fish Fisheries, 2(1):78-85.
McDowall, R. M. (2007). On amphidromy, a distinct form of diadromy in aquatic organisms. Fish Fisheries, 8(1):1-13.
Modeel, S., Joshi, B. D., Yadav, S., Bharti, M., & Negi, R. K. (2023). Mitochondrial DNA reveals shallow population genetic structure in economically important Cyprinid fish Labeo rohita (Hamilton, 1822) from South and Southeast Asia. Molecular Biology Reports, 50(6):4759-4767.
O'Dwyer, J. E., Murphy, N., Tonkin, Z., Lyon, J., Koster, W., Dawson, D., Amtstaetter, F., & Harrisson, K. A. (2021). An investigation of genetic connectivity shines a light on the relative roles of isolation by distance and oceanic currents in three diadromous fish species. Marine and Freshwater Research, 72(10):1457-1473.
Orlova, S. Y., Emelyanova, O. R., Nebesikhina, N. A. Rabazanov, N. I., & Orlov, A. M. (2024). The problems of DNA-barcoding the shads of genus Alosa (Alosidae) of the Ponto-Caspian Basin. Journal of Ichthyology, 64(3):510-520.
Perrier, C., Ferchaud, A-L., Sirois, P., Thibault, I., & Bernatchez, l. (2017). Do genetic drift and accumulation of deleterious mutations preclude adaptation? Empirical investigation using RADseq in a northern lacustrine fish. Molecular Ecology, 26(22):6317-6335.
Peery, M. Z., Kirby, R., Reid, B. N., Stoelting, R., Doucet-Bëer, E., Robinson, S., Vásquez-Carrillo, C., Pauli, J. N., & Palsbøll, P. J. (2012). Reliability of genetic bottleneck tests for detecting recent population declines. Molecular Ecology, 21(14):3403-3418.
Pinacho-Pinacho, C., D., Guzmán-Valdivieso, I., Calixto-Rojas, M., García-Vásquez, A., & Rubio-Godoy, M. (2023). Morphological and molecular characterization of three new species of Gyrodactylus (Monogenea: Gyrodactylidae) infecting Sicydium salvini (Teleostei: Gobiidae) in Mexican rivers draining into the Pacific Ocean. Parasitology Internasional, 93(102712):1-12.
Prabowo, T., Simanjuntak, C. P. H., Affandi, R., Baihaqi, F., Annida, S. B., Ervinia, A., & Budiman, M. S. (2022). Temporal variation in abundance of bully sleepers (Pisces: Eleotridae) larvae and juveniles in Cisolok Estuary, Palabuhanratu Bay, Indonesia. IOP Conference Series: Earth Environmental Science, 1033(012003):1-9.
Ramírez-Soriano, A., Ramos-Onsins, S. E., Rozas, J., Calafell, F., & Navarro, A. (2008). Statistical power analysis of neutrality tests under demographic expansions, contractions and bottlenecks with recombination. Genetics, 179(1):555-567.
Ramos-Onsins, S. E., & Rozas, J. (2006). Statistical properties of new neutrality tests against population growth. Molecular Biology and Evolution, 23(8):2092-2100.
Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., & Sánchez-Gracia, A. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution, 34(12):3299-3302.
Sabatino, S. J., Faria, R., & Alexandrino, P. B. (2022). Genetic structure, diversity, and connectivity in anadromous and freshwater Alosa alosa and A. fallax. Marine Biology, 169(1):1-14.
Sahami, F. M., Kepel, R. C., Olii, A. H., Pratasik, S. B., Lasabuda, R., Wantasen, A., & Habibie, S. A. (2020). Morphometric and genetic variations of species composers of nike fish assemblages in Gorontalo Bay Waters, Indonesia. Biodiversitas, 21(10):4571-4581.
Salmenkova, E., A. (2017). Mechanisms of homing in salmonids. Biology Bulletin Reviews, 7(4):287-298.
Sánchez-Montes, G., Ariño, A. H., Vizmanos, J. L., Wang, J., & Martínez-Solano, Í. (2017). Effects of sample size and full sibs on genetic diversity characterization: A case study of three syntopic iberian pond-breeding amphibians. Journal of Heredity, 108(5):535-543.
Selbie, D. T., Lewis, B. A., Smol, J. P., & Finney, B. P. (2011). Long‐term population dynamics of the endangered snake river sockeye salmon: Evidence of past influences on stock decline and impediments to recovery. Transactions of the American Fisheries Society, 136(3):800-821.
Simanjuntak, C. P. H., Baihaqi, F., Prabowo, T., Bilqis, A. S., Sulistiono, & Ervinia, A. (2021). Recruitment patterns of freshwater amphidromous fishes (Pisces: Gobiidae, Eleotridae) to the Cimaja estuary, Palabuhanratu Bay. [in Indonesian] Jurnal Iktiologi Indonesia, 21(3):321-337.
Taillebois, L., Castelin, M., Ovenden, J. R., Bonillo, C., & Keith, P. (2013). Contrasting genetic structure among populations of two amphidromous fish species (Sicydiinae) in the Central West Pacific. PLoS ONE, 8(10):1-13.
Tamura, K., Stecher, G., & Kumar, S. (2021). MEGA11: Molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution, 38(7):3022-3027.
Walter, R., Hogan, J., Blum, M., Gagne, R., Hain, E., Gilliam, J., & McIntyre, P. (2012). Climate change and conservation of endemic amphidromous fishes in Hawaiian streams. Endangered Species Research, 16:261-272.
Wang, M., Zhang, X., Yang, T., Han, Z., Yanagimoto, T., & Gao, T. (2008). Genetic diversity in the mtDNA control region and population structure in the Sardinella zunasi Bleeker. African Journal of Biotechnology, 7(24):4384-4392.
Watanabe, S., Iida, M., Kimura, Y., Feunteun, E., & Tsukamoto, K. (2006). Genetic diversity of Sicyopterus japonicus as revealed by mitochondrial DNA sequencing. Coastal Marine Science, 30(2):473-479.
Xuan, Z., Jiang, T., Liu, H., Chen, X., & Yang, J. (2021). Mitochondrial DNA and microsatellite analyses reveal strong genetic differentiation between two types of estuarine tapertail anchovies (Coilia) in Yangtze River Basin, China. Hydrobiologia, 848:1409-1431.
Zhao, F., Liu, Y., Wang, Z., Lu, J., Cao, L., & Zeng, C. (2023). Genetic diversity and connectivity of Ocypode ceratophthalmus in the East and South China Seas and its implications for conservation. Biology, 12(437):1-13.